Tutorial - Joint Analysis & Label Transfer
This tutorial describes how to use the Dual Mode in Cellar and related functionality such as Label Transfer. A video tutorial can be found here.
We will be analyzing a SNARE-seq dataset where the two modalities have been split into separate AnnData objects. These datasets can be found on Cellar under the names kidney_SNARE_ATAC_20201005 for the chromatin modality and kidney_SNARE_RNA_20201005 for the expression modality.
- First load the chromatin data by selecting kidney_SNARE_ATAC_20201005 from the dropdown menu. Click
Load.
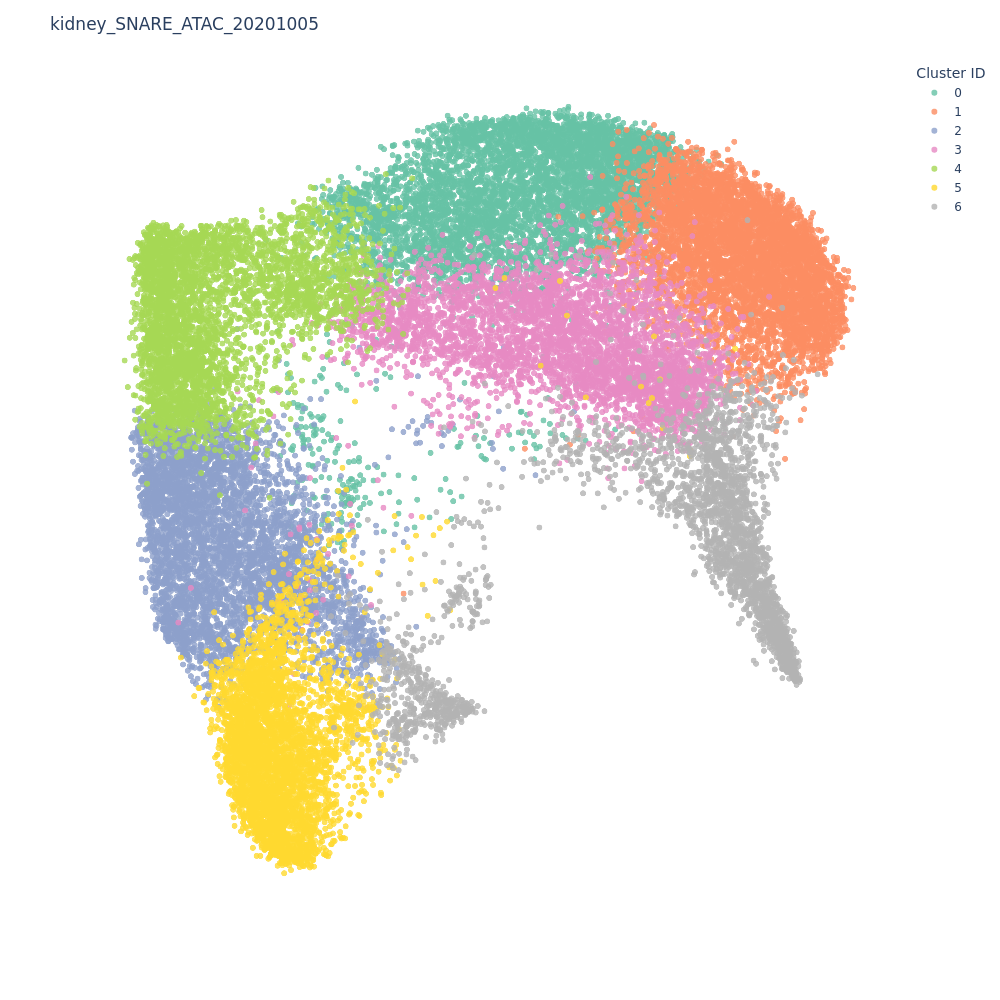
We can see at the top left corner of the app that this dataset contains 31,758 cells and 575,265 features (peaks). This high dimensionality combined with the sparsity makes it very hard to use methods like PCA or UMAP to reduce this data. cisTopic can be a good alternative in this case. cisTopic will find cis-regulatory topics by using a probabilistic model based on Latent Dirichlet allocation, which can be interpreted as a reduced version of the data. Running cisTopic on 500k features might take a long time (up to an hour). However, we have already done this for you and you should already see a 2D representation of the topics on the screen, clustered by Leiden.
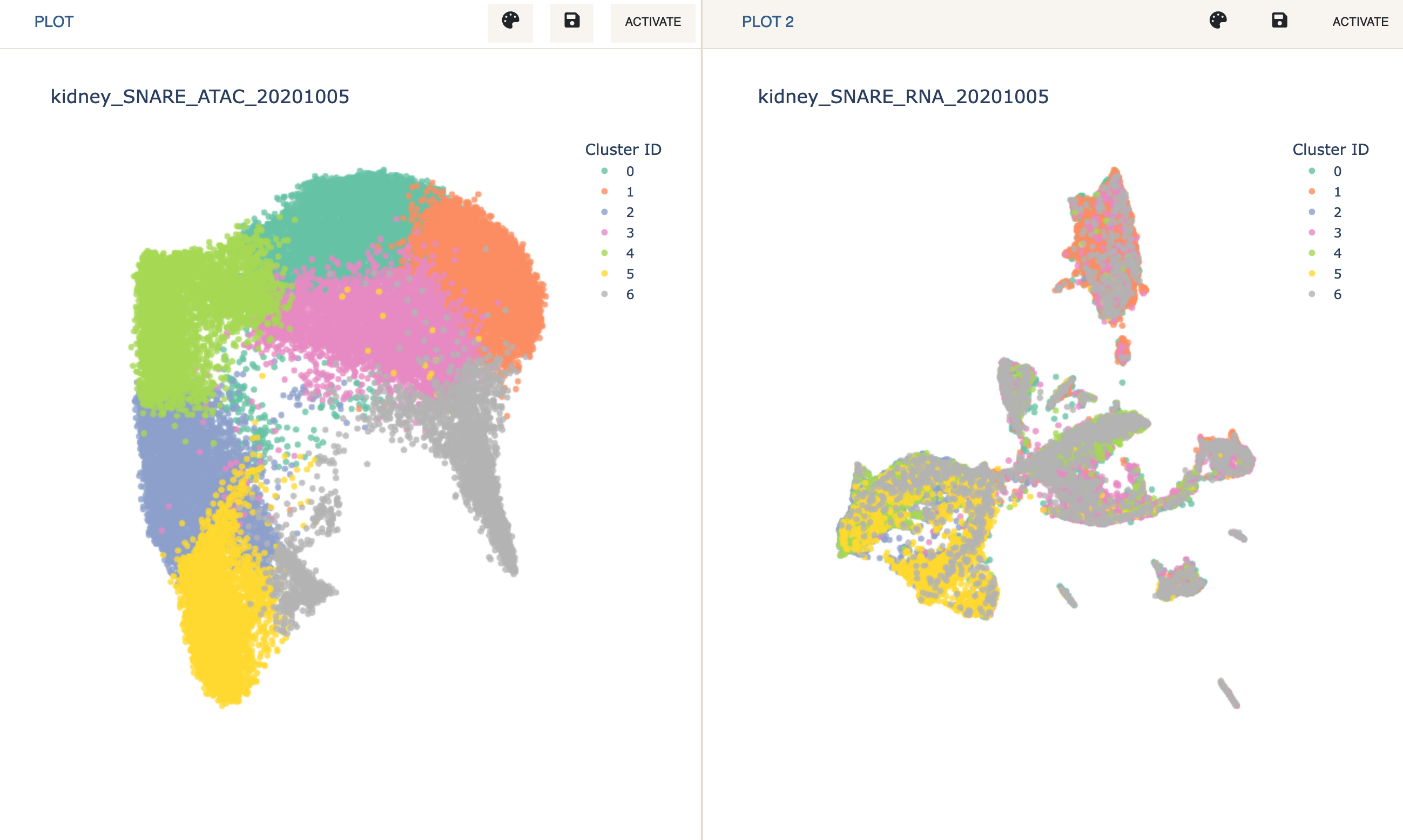
Next, we enter Dual Mode and load the expression data on the 2nd plot.
- Click
Dual Modeon the navigation bar. This will split the screen in two. NowActivatethe empty plot that was just created and load kidney_SNARE_RNA_20201005 just like you normally would.
- Reduce the dimensions of the expression data via PCA + UMAP.
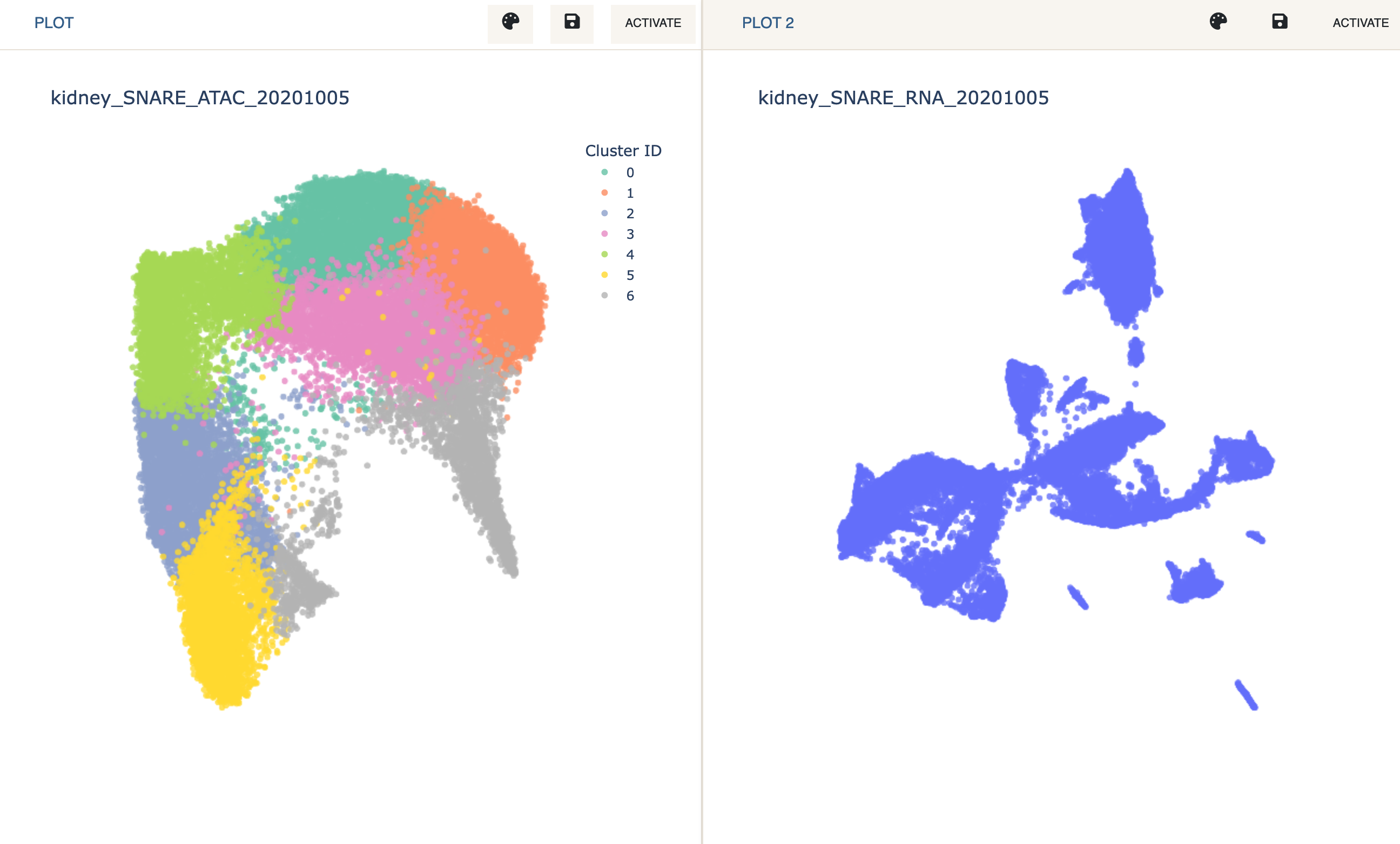
We now wish to transfer labels from one plot to the other. This can be obtained by going to the Label Transfer panel. Since cell IDs between the two datasets match, we can use the Cell ID Based label transfer method.
NOTE: we are overloading the term “label transfer” as no real label transfer is happening here. Actual integration methods include Scanpy Ingest and SingleR. These methods can be used between two datasets that do not necessarily come from the same experiment. The procedure is similar to what follows.
- Under the
Label Transfertab in theClusteringpanel, selectCell ID basedand clickRun.
We can now see the cluster assignments mapped from the first modality to the second.