Tools
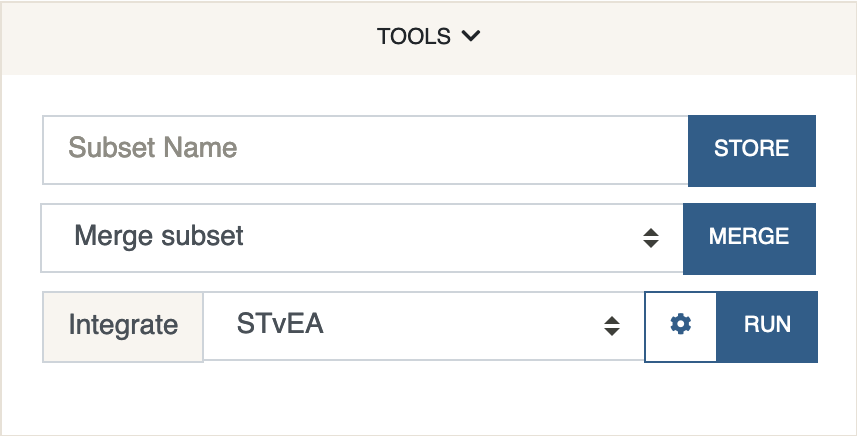
This panel consists of general tools. Currently it is only being used to define new subsets or merge existing ones, and for performing STvEA data integration between CITE-seq and CODEX protein data.
-
Store Subset
To store a subset, a group of cells must be selected from the plot first. This can be done using the lasso tool that is active by default, but can also be switched to anytime in the plot’s toolbox.
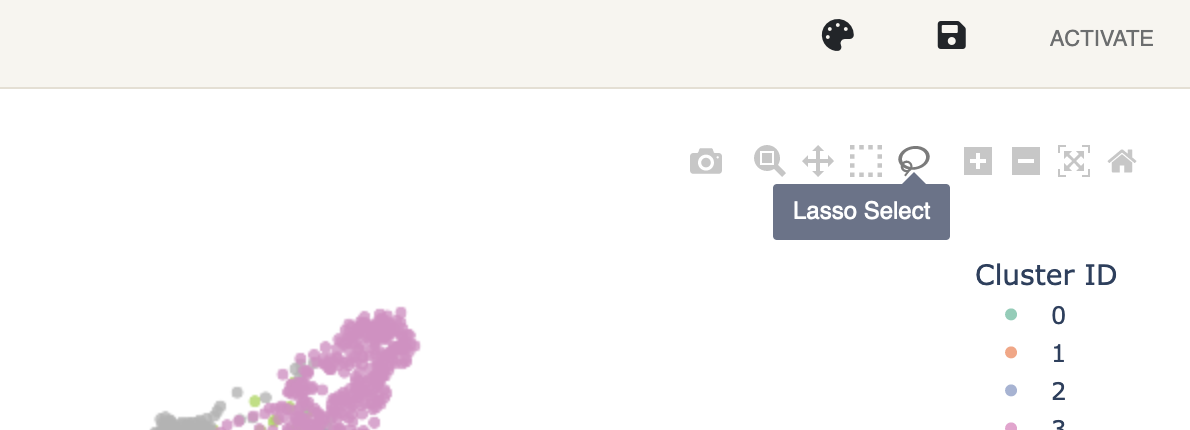 Clicking and dragging your mouse while the lasso tool is active will select and highlight a group of cells which can then be given a name tag. Double clicking on the plot will reset the selection.
Clicking and dragging your mouse while the lasso tool is active will select and highlight a group of cells which can then be given a name tag. Double clicking on the plot will reset the selection. -
Merge Subset
Any subset of cells as defined previously can be merged to form a new cluster. The new cluster will automatically be assigned an ID that is greater than all the cluster IDs before the merge.
-
Integrate
Currently only implements STvEA. STvEA can be used to integrate protein data from CITE-seq and CODEX. More precisely, it is used to map gene expression data from CITE-seq to CODEX. For this method to work, the CITE-seq anndata object must contain protein expression data stored under
adata.obsm['proteins']while the main matrixadata.Xshould hold the gene expression counterpart. Protein names should be stored underadata.uns['proteins']and they must match CODEX protein names stored inadata.var_names. Running STvEA will add the mapped gene expression values underadata.obsm['genes']in the CODEX anndata file. These genes can be selected for visualization under “Other” Features in the Analysis panel.STvEA works best if the protein expression levels are contained in the (0, 1) interval, so we recommend having the cleaning settings set to True.
Subsets are stored under adata.uns['subsets'].